Diagnostic accuracy of deep learning in medical imaging: a systematic review and meta-analysis
Deep learning (DL) has the potential to transform medical diagnostics. However, the diagnostic accuracy of DL is uncertain. Our aim was to evaluate the diagnostic accuracy of DL algorithms to identify pathology in medical imaging. Searches were conducted in Medline and EMBASE up to January 2020. We identified 11,921 studies, of which 503 were included in the systematic review. Eighty-two studies in ophthalmology, 82 in breast disease and 115 in respiratory disease were included for meta-analysis. Two hundred twenty-four studies in other specialities were included for qualitative review. Peer-reviewed studies that reported on the diagnostic accuracy of DL algorithms to identify pathology using medical imaging were included. Primary outcomes were measures of diagnostic accuracy, study design and reporting standards in the literature. Estimates were pooled using random-effects meta-analysis. In ophthalmology, AUC’s ranged between 0.933 and 1 for diagnosing diabetic retinopathy, age-related macular degeneration and glaucoma on retinal fundus photographs and optical coherence tomography. In respiratory imaging, AUC’s ranged between 0.864 and 0.937 for diagnosing lung nodules or lung cancer on chest X-ray or CT scan. For breast imaging, AUC’s ranged between 0.868 and 0.909 for diagnosing breast cancer on mammogram, ultrasound, MRI and digital breast tomosynthesis. Heterogeneity was high between studies and extensive variation in methodology, terminology and outcome measures was noted. This can lead to an overestimation of the diagnostic accuracy of DL algorithms on medical imaging. There is an immediate need for the development of artificial intelligence-specific EQUATOR guidelines, particularly STARD, in order to provide guidance around key issues in this field.
Similar content being viewed by others
Designing deep learning studies in cancer diagnostics
Article 29 January 2021
Machine learning for medical imaging: methodological failures and recommendations for the future
Article Open access 12 April 2022

Artificial intelligence in digital pathology: a systematic review and meta-analysis of diagnostic test accuracy
Article Open access 04 May 2024
Introduction
Artificial Intelligence (AI), and its subfield of deep learning (DL) 1 , offers the prospect of descriptive, predictive and prescriptive analysis, in order to attain insight that would otherwise be untenable through manual analyses 2 . DL-based algorithms, using architectures such as convolutional neural networks (CNNs), are distinct from traditional machine learning approaches. They are distinguished by their ability to learn complex representations in order to improve pattern recognition from raw data, rather than requiring human engineering and domain expertise to structure data and design feature extractors 3 .
Of all avenues through which DL may be applied to healthcare; medical imaging, part of the wider remit of diagnostics, is seen as the largest and most promising field 4,5 . Currently, radiological investigations, regardless of modality, require interpretation by a human radiologist in order to attain a diagnosis in a timely fashion. With increasing demands upon existing radiologists (especially in low-to-middle-income countries) 6,7,8 , there is a growing need for diagnosis automation. This is an issue that DL is able to address 9 .
Successful integration of DL technology into routine clinical practice relies upon achieving diagnostic accuracy that is non-inferior to healthcare professionals. In addition, it must provide other benefits, such as speed, efficiency, cost, bolstering accessibility and the maintenance of ethical conduct.
Although regulatory approval has already been granted by the Food and Drug Administration for select DL-powered diagnostic software to be used in clinical practice 10,11 , many note that the critical appraisal and independent evaluation of these technologies are still in their infancy 12 . Even within seminal studies in the field, there remains wide variation in design, methodology and reporting that limits the generalisability and applicability of their findings 13 . Moreover, it is noted that there has been no overarching medical specialty-specific meta-analysis assessing diagnostic accuracy of DL performance, particularly in ophthalmology, respiratory medicine and breast surgery, which have the most diagnostic studies to date 13 .
Therefore, the aim of this review is to (1) quantify the diagnostic accuracy of DL in speciality-specific radiological imaging modalities to identify or classify disease, and (2) to appraise the variation in methodology and reporting of DL-based radiological diagnosis, in order to highlight the most common flaws that are pervasive across the field.
Results
Search and study selection
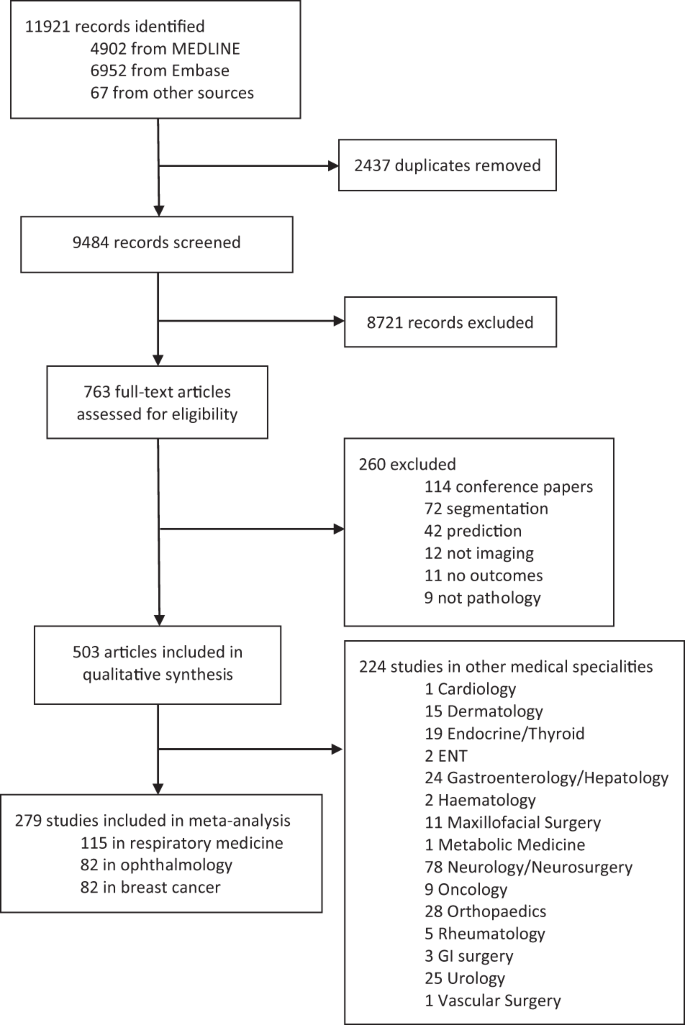
Our search identified 11,921 abstracts, of which 9484 were screened after duplicates were removed. Of these, 8721 did not fulfil inclusion criteria based on title and abstract. Seven hundred sixty-three full manuscripts were individually assessed and 260 were excluded at this step. Five hundred three papers fulfilled inclusion criteria for the systematic review and contained data required for sensitivity, specificity or AUC. Two hundred seventy-three studies were included for meta-analysis, 82 in ophthalmology, 115 in respiratory medicine and 82 in breast cancer (see Fig. 1). These three fields were chosen to meta-analyse as they had the largest numbers of studies with available data. Two hundred twenty-four other studies were included for qualitative synthesis in other medical specialities. Summary estimates of imaging and speciality-specific diagnostic accuracy metrics are described in Table 1. Units of analysis for each speciality and modality are indicated in Tables 2–4.
Discussion
This study sought to (1) quantify the diagnostic accuracy of DL algorithms to identify specific pathology across distinct radiological modalities, and (2) appraise the variation in study reporting of DL-based radiological diagnosis. The findings of our speciality-specific meta-analysis suggest that DL algorithms generally have a high and clinically acceptable diagnostic accuracy in identifying disease. High diagnostic accuracy with analogous DL approaches was identified in all specialities despite different workflows, pathology and imaging modalities, suggesting that DL algorithms can be deployed across different areas in radiology. However, due to high heterogeneity and variance between studies, there is considerable uncertainty around estimates of diagnostic accuracy in this meta-analysis.
In ophthalmology, the findings suggest features of diseases, such as DR, AMD and glaucoma can be identified with a high sensitivity, specificity and AUC, using DL on both RFP and OCT scans. In general, we found higher sensitivity, specificity, accuracy and AUC with DL on OCT scans over RFP for DR, AMD and glaucoma. Only sensitivity was higher for DR on RFP over OCT.
In respiratory medicine, our findings suggest that DL has high sensitivity, specificity and AUC to identify chest pathology on CT scans and CXR. DL on CT had higher sensitivity and AUC for detecting lung nodules; however, we found a higher specificity, PPV and F1 score on CXR. For diagnosing cancer or lung mass, DL on CT had a higher sensitivity than CXR.
In breast cancer imaging, our findings suggest that DL generally has a high diagnostic accuracy to identify breast cancer on mammograms, ultrasound and DBT. The performance was found to be very similar for these modalities. In MRI, however, the diagnostic accuracy was lower; this may be due to small datasets and the use of 2D images. The utilisation of larger databases and multiparametric MRI may increase the diagnostic accuracy 113 .
Extensive variation in the methodology, data interpretability, terminology and outcome measures could be explained by a lack of consensus in how to conduct and report DL studies. The STARD-2015 checklist 114 , designed for reporting of diagnostic accuracy studies is not fully applicable to clinical DL studies 115 . The variation in reporting makes it very difficult to formally evaluate the performance of algorithms. Furthermore, differences in reference standards, grader capabilities, disease definitions and thresholds for diagnosis make direct comparison between studies and algorithms very difficult. This can only be improved with well-designed and executed studies that explicitly address questions concerning transparency, reproducibility, ethics and effectiveness 116 and specific reporting standards for AI studies 115,117 .
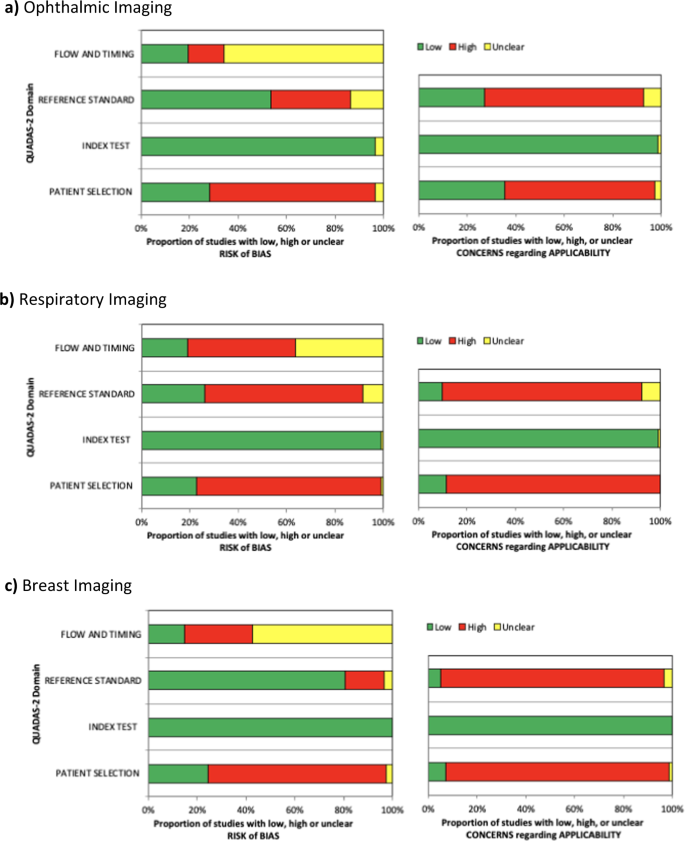
The QUADAS-2 (ref. 118 ) assessment tool was used to systematically evaluate the risk of bias and any applicability concerns of the diagnostic accuracy studies. Although this tool was not designed for DL diagnostic accuracy studies, the evaluation allowed us to judge that a majority of studies in this field are at risk of bias or concerning for applicability. Of particular concern was the applicability of reference standards and patient selection.
Despite our results demonstrating that DL algorithms have a high diagnostic accuracy in medical imaging, it is currently difficult to determine if they are clinically acceptable or applicable. This is partially due to the extensive variation and risk of bias identified in the literature to date. Furthermore, the definition of what threshold is acceptable for clinical use and tolerance for errors varies greatly across diseases and clinical scenarios 119 .
Limitations in the literature
Dataset
There are broad methodological deficiencies among the included studies. Most studies were performed using retrospectively collected data, using reference standards and labels that were not intended for the purposes of DL analysis. Minimal prospective studies and only two randomised studies 109,120 , evaluating the performance of DL algorithms in clinical settings were identified in the literature. Proper acquisition of test data is essential to interpret model performance in a real-world clinical setting. Poor quality reference standards may result in the decreased model performance due to suboptimal data labelling in the validation set 28 , which could be a barrier to understanding the true capabilities of the model on the test set. This is symptomatic of the larger issue that there is a paucity of gold-standard, prospectively collected, representative datasets for the purposes of DL model testing. However, as there are many advantages to using retrospectively collected data, the resourceful use of retrospective or synthetic data with the use of labels of varying modality and quality represent important areas of research in DL 121 .
Study methodology
Many studies did not undertake external validation of the algorithm in a separate test set and relied upon results from the internal validation data; the same dataset used to train the algorithm initially. This may lead to an overestimation of the diagnostic accuracy of the algorithm. The problem of overfitting has been well described in relation to machine learning algorithms 122 . True demonstration of the performance of these algorithms can only be assumed if they are externally validated on separate test sets with previously unseen data that are representative of the target population.
Surprisingly, few studies compared the diagnostic accuracy of DL algorithms against expert human clinicians for medical imaging. This would provide a more objective standard that would enable better comparison of models across studies. Furthermore, application of the same test dataset for diagnostic performance assessment of DL algorithms versus healthcare professionals was identified in only select studies 13 . This methodological deficiency limits the ability to gauge the clinical applicability of these algorithms into clinical practice. Similarly, this issue can extend to model-versus-model comparisons. Specific methods of model training or model architecture may not be described well enough to permit emulation for comparison 123 . Thus, standards for model development and comparison against controls will be needed as DL architectures and techniques continue to develop and are applied in medical contexts.
Reporting
There was varying terminology and a lack of transparency used in DL studies with regards to the validation or test sets used. The term ‘validation’ was identified as being used interchangeably to either describe an external test set for the final algorithm or for an internal dataset that is used to fine tune the model prior to ‘testing’. Furthermore, the inconsistent terminology led to difficulties understanding whether an independent external test set was used to test diagnostic performance 13 .
Crucially, we found broad variation in the metrics used as outcomes for the performance of the DL algorithms in the literature. Very few studies reported true positives, false positives, true negatives and false negatives in a contingency table as should be the minimum for diagnostic accuracy studies 114 . Moreover, some studies only reported metrics, such as dice coefficient, F1 score, competition performance metric and Top-1 accuracy that are often used in computer science, but may be unfamiliar to clinicians 13 . Metrics such as AUC, sensitivity, specificity, PPV and NPV should be reported, as these are more widely understood by healthcare professionals. However, it is noted that NPV and PPV are dependent on the underlying prevalence of disease and as many test sets are artificially constructed or balanced, then reporting the NPV or PPV may not be valid. The wide range of metrics reported also leads to difficulty in comparing the performance of algorithms on similar datasets.
Study strengths and limitations
This systematic review and meta-analysis statistically appraises pooled data collected from 279 studies. It is the largest study to date examining the diagnostic accuracy of DL on medical imaging. However, our findings must be viewed in consideration of several limitations. Firstly, as we believe that many studies have methodological deficiencies or are poorly reported, these studies may not be a reliable source for evaluating diagnostic accuracy. Consequently, the estimates of diagnostic performance provided in our meta-analysis are uncertain and may represent an over-estimation of the true accuracy. Secondly, we did not conduct a quality assessment for the transparency of reporting in this review. This was because current guidelines to assess diagnostic accuracy reporting standards (STARD-2015 114 ) were not designed for DL studies and are not fully applicable to the specifics and nuances of DL research 115 . Thirdly, due to the nature of DL studies, we were not able to perform classical statistical comparison of measures of diagnostic accuracy between different imaging modalities. Fourthly, we were unable to separate each imaging modality into different subsets, to enable comparison across subsets and allow the heterogeneity and variance to be broken down. This was because our study aimed to provide an overview of the literature in each specific speciality, and it was beyond the scope of this review to examine each modality individually. The inherent differences in imaging technology, patient populations, pathologies and study designs meant that attempting to derive common lessons across the board did not always offer easy comparisons. Finally, our review concentrated on DL for speciality-specific medical imaging, and therefore it may not be appropriate to generalise our findings to other forms of medical imaging or AI studies.
Future work
For the quality of DL research to flourish in the future, we believe that the adoption of the following recommendations are required as a starting point.
Availability of large, open-source, diverse anonymised datasets with annotations
This can be achieved through governmental support and will enable greater reproducibility of DL models 124 .
Collaboration with academic centres to utilise their expertise in pragmatic trial design and methodology 125
Rather than classical trials, novel experimental and quasi-experimental methods to evaluate DL have been proposed and should be evaluated 126 . This may include ongoing evaluation of algorithms once in clinical practice, as they continue to learn and adapt to the population that they are implemented in.
Creation of AI-specific reporting standards
A major reason for the difficulties encountered in evaluating the performance of DL on medical imaging are largely due to inconsistent and haphazard reporting. Although DL is widely considered as a ‘predictive’ model (where TRIPOD may be applied) the majority of AI interventions close to translation currently published are predominantly in the field of diagnostics (with specifics on index tests, reference standards and true/false positive/negatives and summary diagnostic scores, centred directly in the domain of STARD). Existing reporting guidelines for diagnostic accuracy studies (STARD) 114 , prediction models (TRIPOD) 127 , randomised trials (CONSORT) 128 and interventional trial protocols (SPIRIT) 129 do not fully cover DL research due to specific considerations in methodology, data and interpretation required for these studies. As such, we applaud the recent publication of the CONSORT-AI 117 and SPIRIT-AI 130 guidelines, and await AI-specific amendments of the TRIPOD-AI 131 and STARD-AI 115 statements (which we are convening). We trust that when these are published, studies being conducted will have a framework that enables higher quality and more consistent reporting.
Development of specific tools for determining the risk of study bias and applicability
An update to the QUADAS-2 tool taking into account the nuances of DL diagnostic accuracy research should be considered.
Updated specific ethical and legal framework
Outdated policies need to be updated and key questions answered in terms of liability in cases of medical error, doctor and patient understanding, control over algorithms and protection of medical data 132 . The World Health Organisation 133 and others have started to develop guidelines and principles to regulate the use of AI. These regulations will need to be adapted by each country to fit their own political and healthcare context 134 . Furthermore, these guidelines will need to proactively and objectively evaluate technology to ensure best practices are developed and implemented in an evidence-based manner 135 .
Conclusion
DL is a rapidly developing field that has great potential in all aspects of healthcare, particularly radiology. This systematic review and meta-analysis appraised the quality of the literature and provided pooled diagnostic accuracy for DL techniques in three medical specialities. While the results demonstrate that DL currently has a high diagnostic accuracy, it is important that these findings are assumed in the presence of poor design, conduct and reporting of studies, which can lead to bias and overestimating the power of these algorithms. The application of DL can only be improved with standardised guidance around study design and reporting, which could help clarify clinical utility in the future. There is an immediate need for the development of AI-specific STARD and TRIPOD statements to provide robust guidance around key issues in this field before the potential of DL in diagnostic healthcare is truly realised in clinical practice.
Methods
This systematic review was conducted in accordance with the guidelines for the ‘Preferred Reporting Items for Systematic Reviews and Meta-Analyses’ extension for diagnostic accuracy studies statement (PRISMA-DTA) 136 .
Eligibility criteria
Studies that report upon the diagnostic accuracy of DL algorithms to investigate pathology or disease on medical imaging were sought. The primary outcome was various diagnostic accuracy metrics. Secondary outcomes were study design and quality of reporting.
Data sources and searches
Electronic bibliographic searches were conducted in Medline and EMBASE up to 3rd January 2020. MESH terms and all-field search terms were searched for ‘neural networks’ (DL or convolutional or cnn) and ‘imaging’ (magnetic resonance or computed tomography or OCT or ultrasound or X-ray) and ‘diagnostic accuracy metrics’ (sensitivity or specificity or AUC). For the full search strategy, please see Supplementary Methods 1. The search included all study designs. Further studies were identified through manual searches of bibliographies and citations until no further relevant studies were identified. Two investigators (R.A. and V.S.) independently screened titles and abstracts, and selected all relevant citations for full-text review. Disagreement regarding study inclusion was resolved by discussion with a third investigator (H.A.).
Inclusion criteria
Studies that comprised a diagnostic accuracy assessment of a DL algorithm on medical imaging in human populations were eligible. Only studies that stated either diagnostic accuracy raw data, or sensitivity, specificity, AUC, NPV, PPV or accuracy data were included in the meta-analysis. No limitations were placed on the date range and the last search was performed in January 2020.
Exclusion criteria
Articles were excluded if the article was not written in English. Abstracts, conference articles, pre-prints, reviews and meta-analyses were not considered because an aim of this review was to appraise the methodology, reporting standards and quality of primary research studies being published in peer-reviewed journals. Studies that investigated the accuracy of image segmentation or predicting disease rather than identification or classification were excluded.
Data extraction and quality assessment
Two investigators (R.A. and V.S.) independently extracted demographic and diagnostic accuracy data from the studies, using a predefined electronic data extraction spreadsheet. The data fields were chosen subsequent to an initial scoping review and were, in the opinion of the investigators, sufficient to fulfil the aims of this review. Data were extracted on (i) first author, (ii) year of publication, (iii) type of neural network, (iv) population, (v) dataset—split into training, validation and test sets, (vi) imaging modality, (vii) body system/disease, (viii) internal/external validation methods, (ix) reference standard, (x) diagnostic accuracy raw data—true and false positives and negatives, (xi) percentages of AUC, accuracy, sensitivity, specificity, PPV, NPV and other metrics reported.
Three investigators (R.A., V.S. and GM) assessed study methodology using the QUADAS-2 checklist to evaluate the risk of bias and any applicability concerns of the studies 118 .
Data synthesis and analysis
A bivariate model for diagnostic meta-analysis was used to calculate summary estimates of sensitivity, specificity and AUC data 137 . Independent proportion and their differences were calculated and pooled through DerSimonian and Laird random-effects modelling 138 . This considered both between-study and within-study variances that contributed to study weighting. Study-specific estimates and 95% CIs were computed and represented on forest plots. Heterogeneity between studies was assessed using I 2 (25–49% was considered to be low heterogeneity, 50–74% was moderate and >75% was high heterogeneity). Where raw diagnostic accuracy data were available, the SROC model was used to evaluate the relationship between sensitivity and specificity 139 . We utilised Stata version 15 (Stata Corp LP, College Station, TX, USA) for all statistical analyses.
We chose to appraise the performance of DL algorithms to identify individual disease or pathology patterns on different imaging modalities in isolation, e.g., identifying lung nodules on a thoracic CT scan. We felt that combining imaging modalities and diagnoses would add heterogeneity and variation to the analysis. Meta-analysis was only performed where there were greater than or equal to three patient cohorts, reporting for each specific pathology and imaging modality. This study is registered with PROSPERO, CRD42020167503.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The authors declare that all the data included in this study are available within the paper and its Supplementary Information files.
References
- LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature521, 436–444 (2015). ArticleCASPubMedGoogle Scholar
- Obermeyer, Z. & Emanuel, E. J. Predicting the future — big data, machine learning, and clinical medicine. N. Engl. J. Med.375, 1216–1219 (2016). ArticlePubMedPubMed CentralGoogle Scholar
- Esteva, A. et al. A guide to deep learning in healthcare. Nat. Med.25, 24–29 (2019). ArticleCASPubMedGoogle Scholar
- Litjens, G. et al. A survey on deep learning in medical image analysis. Med. Image Anal.42, 60–88 (2017). ArticlePubMedGoogle Scholar
- Bluemke, D. A. et al. Assessing radiology research on artificial intelligence: a brief guide for authors, reviewers, and readers—from the radiology editorial board. Radiology294, 487–489 (2020). ArticlePubMedGoogle Scholar
- Wahl, B., Cossy-Gantner, A., Germann, S. & Schwalbe, N. R. Artificial intelligence (AI) and global health: how can AI contribute to health in resource-poor settings? BMJ Glob. Health3, e000798–e000798 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Zhang, L., Wang, H., Li, Q., Zhao, M.-H. & Zhan, Q.-M. Big data and medical research in China. BMJ360, j5910 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Nakajima, Y., Yamada, K., Imamura, K. & Kobayashi, K. Radiologist supply and workload: international comparison. Radiat. Med.26, 455–465 (2008). ArticlePubMedGoogle Scholar
- Kelly, C. J., Karthikesalingam, A., Suleyman, M., Corrado, G. & King, D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med.17, 195 (2019). ArticlePubMedPubMed CentralCASGoogle Scholar
- Topol, E. J. High-performance medicine: the convergence of human and artificial intelligence. Nat. Med.25, 44–56 (2019). ArticleCASPubMedGoogle Scholar
- Benjamens, S., Dhunnoo, P. & Meskó, B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: an online database. npj Digital Med.3, 118 (2020). ArticleGoogle Scholar
- Beam, A. L. & Kohane, I. S. Big data and machine learning in health care. JAMA319, 1317–1318 (2018). ArticlePubMedGoogle Scholar
- Liu, X. et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digital Health1, e271–e297 (2019). ArticlePubMedGoogle Scholar
- Abràmoff, M. D., Lavin, P. T., Birch, M., Shah, N. & Folk, J. C. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. npj Digital Med.1, 39 (2018). ArticleGoogle Scholar
- Bellemo, V. et al. Artificial intelligence using deep learning to screen for referable and vision-threatening diabetic retinopathy in Africa: a clinical validation study. Lancet Digital Health1, e35–e44 (2019). ArticlePubMedGoogle Scholar
- Christopher, M. et al. Performance of deep learning architectures and transfer learning for detecting glaucomatous optic neuropathy in fundus photographs. Sci. Rep.8, 16685 (2018). ArticlePubMedPubMed CentralCASGoogle Scholar
- Gulshan, V. et al. Performance of a deep-learning algorithm vs manual grading for detecting diabetic retinopathy in India. JAMA Ophthalmol137, 987–993 (2019). ArticlePubMed CentralPubMedGoogle Scholar
- Keel, S., Wu, J., Lee, P. Y., Scheetz, J. & He, M. Visualizing deep learning models for the detection of referable diabetic retinopathy and glaucoma. JAMA Ophthalmol.137, 288–292 (2019). ArticlePubMedGoogle Scholar
- Sandhu, H. S. et al. Automated diagnosis and grading of diabetic retinopathy using optical coherence tomography. Investig. Ophthalmol. Vis. Sci.59, 3155–3160 (2018). ArticleGoogle Scholar
- Zheng, C. et al. Detecting glaucoma based on spectral domain optical coherence tomography imaging of peripapillary retinal nerve fiber layer: a comparison study between hand-crafted features and deep learning model. Graefes Arch. Clin. Exp. Ophthalmol.258, 577–585 (2020). ArticlePubMedGoogle Scholar
- Kanagasingam, Y. et al. Evaluation of artificial intelligence-based grading of diabetic retinopathy in primary care. JAMA Netw. Open1, e182665–e182665 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Alqudah, A. M. AOCT-NET: a convolutional network automated classification of multiclass retinal diseases using spectral-domain optical coherence tomography images. Med. Biol. Eng. Comput.58, 41–53 (2020). ArticlePubMedGoogle Scholar
- Asaoka, R. et al. Validation of a deep learning model to screen for glaucoma using images from different fundus cameras and data augmentation. Ophthalmol. Glaucoma2, 224–231 (2019). ArticlePubMedGoogle Scholar
- Bhatia, K. K. et al. Disease classification of macular optical coherence tomography scans using deep learning software: validation on independent, multicenter data. Retina40, 1549–1557 (2020). ArticlePubMedGoogle Scholar
- Chan, G. C. Y. et al. Fusing results of several deep learning architectures for automatic classification of normal and diabetic macular edema in optical coherence tomography. In Conference proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual Conference, Vol. 2018, 670–673 (IEEE, 2018).
- Gargeya, R. & Leng, T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology124, 962–969 (2017). ArticlePubMedGoogle Scholar
- Grassmann, F. et al. A deep learning algorithm for prediction of age-related eye disease study severity scale for age-related macular degeneration from color fundus photography. Ophthalmology125, 1410–1420 (2018). ArticlePubMedGoogle Scholar
- Gulshan, V. et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA316, 2402–2410 (2016). ArticlePubMedGoogle Scholar
- Hwang, D. K. et al. Artificial intelligence-based decision-making for age-related macular degeneration. Theranostics9, 232–245 (2019). ArticlePubMedPubMed CentralGoogle Scholar
- Keel, S. et al. Development and validation of a deep-learning algorithm for the detection of neovascular age-related macular degeneration from colour fundus photographs. Clin. Exp. Ophthalmol.47, 1009–1018 (2019). ArticlePubMedGoogle Scholar
- Krause, J. et al. Grader variability and the importance of reference standards for evaluating machine learning models for diabetic retinopathy. Ophthalmology125, 1264–1272 (2018). ArticlePubMedGoogle Scholar
- Li, F. et al. Automatic detection of diabetic retinopathy in retinal fundus photographs based on deep learning algorithm. Transl. Vis. Sci. Technol.8, 4 (2019). ArticlePubMedPubMed CentralGoogle Scholar
- Li, Z. et al. An automated grading system for detection of vision-threatening referable diabetic retinopathy on the basis of color fundus photographs. Diabetes Care41, 2509–2516 (2018). ArticlePubMedGoogle Scholar
- Liu, H. et al. Development and validation of a deep learning system to detect glaucomatous optic neuropathy using fundus photographs. JAMA Ophthalmol.137, 1353–1360 (2019). ArticlePubMed CentralPubMedGoogle Scholar
- Liu, S. et al. A deep learning-based algorithm identifies glaucomatous discs using monoscopic fundus photographs. Ophthalmol. Glaucoma1, 15–22 (2018). ArticlePubMedGoogle Scholar
- MacCormick, I. J. C. et al. Accurate, fast, data efficient and interpretable glaucoma diagnosis with automated spatial analysis of the whole cup to disc profile. PLoS ONE14, e0209409 (2019). ArticleCASPubMedPubMed CentralGoogle Scholar
- Phene, S. et al. Deep learning and glaucoma specialists: the relative importance of optic disc features to predict glaucoma referral in fundus photographs. Ophthalmology126, 1627–1639 (2019). ArticlePubMedGoogle Scholar
- Ramachandran, N., Hong, S. C., Sime, M. J. & Wilson, G. A. Diabetic retinopathy screening using deep neural network. Clin. Exp. Ophthalmol.46, 412–416 (2018). ArticlePubMedGoogle Scholar
- Raumviboonsuk, P. et al. Deep learning versus human graders for classifying diabetic retinopathy severity in a nationwide screening program. npj Digital Med.2, 25 (2019). ArticleGoogle Scholar
- Sayres, R. et al. Using a deep learning algorithm and integrated gradients explanation to assist grading for diabetic retinopathy. Ophthalmology126, 552–564 (2019). ArticlePubMedGoogle Scholar
- Ting, D. S. W. et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA318, 2211–2223 (2017). ArticlePubMedPubMed CentralGoogle Scholar
- Ting, D. S. W. et al. Deep learning in estimating prevalence and systemic risk factors for diabetic retinopathy: a multi-ethnic study. npj Digital Med.2, 24 (2019). ArticleGoogle Scholar
- Verbraak, F. D. et al. Diagnostic accuracy of a device for the automated detection of diabetic retinopathy in a primary care setting. Diabetes Care42, 651 (2019). ArticlePubMedGoogle Scholar
- Van Grinsven, M. J., van Ginneken, B., Hoyng, C. B., Theelen, T. & Sánchez, C. I. Fast convolutional neural network training using selective data sampling: application to hemorrhage detection in color fundus images. IEEE Trans. Med. Imaging35, 1273–1284 (2016). ArticlePubMedGoogle Scholar
- Rogers, T. W. et al. Evaluation of an AI system for the automated detection of glaucoma from stereoscopic optic disc photographs: the European Optic Disc Assessment Study. Eye33, 1791–1797 (2019). ArticlePubMedPubMed CentralGoogle Scholar
- Al-Aswad, L. A. et al. Evaluation of a deep learning system for identifying glaucomatous optic neuropathy based on color fundus photographs. J. Glaucoma28, 1029–1034 (2019). ArticlePubMedGoogle Scholar
- Brown, J. M. et al. Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. JAMA Ophthalmol.136, 803–810 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Burlina, P. et al. Utility of deep learning methods for referability classification of age-related macular degeneration. JAMA Ophthalmol.136, 1305–1307 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Burlina, P. M. et al. Automated grading of age-related macular degeneration from color fundus images using deep convolutional neural networks. JAMA Ophthalmol.135, 1170–1176 (2017). ArticlePubMedPubMed CentralGoogle Scholar
- Burlina, P., Pacheco, K. D., Joshi, N., Freund, D. E. & Bressler, N. M. Comparing humans and deep learning performance for grading AMD: a study in using universal deep features and transfer learning for automated AMD analysis. Computers Biol. Med.82, 80–86 (2017). ArticleGoogle Scholar
- De Fauw, J. et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med.24, 1342–1350 (2018). ArticlePubMedCASGoogle Scholar
- Gómez-Valverde, J. J. et al. Automatic glaucoma classification using color fundus images based on convolutional neural networks and transfer learning. Biomed. Opt. Express10, 892–913 (2019). ArticlePubMedPubMed CentralGoogle Scholar
- Jammal, A. A. et al. Human versus machine: comparing a deep learning algorithm to human gradings for detecting glaucoma on fundus photographs. Am. J. Ophthalmol.211, 123–131 (2019). ArticlePubMedPubMed CentralGoogle Scholar
- Kermany, D. S. et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell172, 1122–1131.e1129 (2018). ArticleCASPubMedGoogle Scholar
- Li, F. et al. Deep learning-based automated detection of retinal diseases using optical coherence tomography images. Biomed. Opt. Express10, 6204–6226 (2019). ArticlePubMedPubMed CentralGoogle Scholar
- Long, E. et al. An artificial intelligence platform for the multihospital collaborative management of congenital cataracts. Nat. Biomed. Eng.1, 0024 (2017). ArticleGoogle Scholar
- Matsuba, S. et al. Accuracy of ultra-wide-field fundus ophthalmoscopy-assisted deep learning, a machine-learning technology, for detecting age-related macular degeneration. Int. Ophthalmol.39, 1269–1275 (2019). ArticlePubMedGoogle Scholar
- Nagasato, D. et al. Automated detection of a nonperfusion area caused by retinal vein occlusion in optical coherence tomography angiography images using deep learning. PLoS ONE14, e0223965 (2019). ArticleCASPubMedPubMed CentralGoogle Scholar
- Peng, Y. et al. DeepSeeNet: a deep learning model for automated classification of patient-based age-related macular degeneration severity from color fundus photographs. Ophthalmology126, 565–575 (2019). ArticlePubMedGoogle Scholar
- Shibata, N. et al. Development of a deep residual learning algorithm to screen for glaucoma from fundus photography. Sci. Rep.8, 14665 (2018). ArticlePubMedPubMed CentralCASGoogle Scholar
- Zhang, Y. et al. Development of an automated screening system for retinopathy of prematurity using a deep neural network for wide-angle retinal images. IEEE Access7, 10232–10241 (2019). ArticleGoogle Scholar
- Becker, A. S. et al. Classification of breast cancer in ultrasound imaging using a generic deep learning analysis software: a pilot study. Br. J. Radio.91, 20170576 (2018). Google Scholar
- Zhang, C. et al. Toward an expert level of lung cancer detection and classification using a deep convolutional neural network. Oncologist24, 1159–1165 (2019). ArticlePubMedPubMed CentralGoogle Scholar
- Ardila, D. et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med.25, 954–961 (2019). ArticleCASPubMedGoogle Scholar
- Hwang, E. J. et al. Deep learning for chest radiograph diagnosis in the emergency department. Radiology293, 573–580 (2019). ArticlePubMedGoogle Scholar
- Hwang, E. J. et al. Development and validation of a deep learning–based automated detection algorithm for major thoracic diseases on chest radiographs. JAMA Netw. Open2, e191095–e191095 (2019). ArticlePubMedPubMed CentralGoogle Scholar
- Hwang, E. J. et al. Development and validation of a deep learning–based automatic detection algorithm for active pulmonary tuberculosis on chest radiographs. Clin. Infect. Dis.https://doi.org/10.1093/cid/ciy967 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Liang, C. H. et al. Identifying pulmonary nodules or masses on chest radiography using deep learning: external validation and strategies to improve clinical practice. Clin. Radiol.75, 38–45 (2020). ArticlePubMedGoogle Scholar
- Nam, J. G. et al. Development and validation of deep learning–based automatic detection algorithm for malignant pulmonary nodules on chest radiographs. Radiology290, 218–228 (2018). ArticlePubMedGoogle Scholar
- Qin, Z. Z. et al. Using artificial intelligence to read chest radiographs for tuberculosis detection: A multi-site evaluation of the diagnostic accuracy of three deep learning systems. Sci. Rep.9, 15000 (2019). ArticlePubMedPubMed CentralCASGoogle Scholar
- Setio, A. A. A. et al. Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans. Med. Imaging35, 1160–1169 (2016). ArticlePubMedGoogle Scholar
- Sim, Y. et al. Deep convolutional neural network–based software improves radiologist detection of malignant lung nodules on chest radiographs. Radiology294, 199–209 (2020). ArticlePubMedGoogle Scholar
- Taylor, A. G., Mielke, C. & Mongan, J. Automated detection of moderate and large pneumothorax on frontal chest X-rays using deep convolutional neural networks: a retrospective study. PLOS Med.15, e1002697 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Uthoff, J. et al. Machine learning approach for distinguishing malignant and benign lung nodules utilizing standardized perinodular parenchymal features from CT. Med. Phys.46, 3207–3216 (2019).
- Zech, J. R. et al. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLOS Med.15, e1002683 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Cha, M. J., Chung, M. J., Lee, J. H. & Lee, K. S. Performance of deep learning model in detecting operable lung cancer with chest radiographs. J. Thorac. Imaging34, 86–91 (2019). ArticlePubMedGoogle Scholar
- Chae, K. J. et al. Deep learning for the classification of small (≤2 cm) pulmonary nodules on ct imaging: a preliminary study. Acad. Radiol.27, E55–E63 (2020). ArticlePubMedGoogle Scholar
- Ciompi, F. et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci. Rep.7, 46479 (2017). ArticleCASPubMedPubMed CentralGoogle Scholar
- Dunnmon, J. A. et al. Assessment of convolutional neural networks for automated classification of chest radiographs. Radiology290, 537–544 (2018). ArticlePubMedGoogle Scholar
- Li, X. et al. Deep learning-enabled system for rapid pneumothorax screening on chest CT. Eur. J. Radiol.120, 108692 (2019). ArticlePubMedGoogle Scholar
- Li, L., Liu, Z., Huang, H., Lin, M. & Luo, D. Evaluating the performance of a deep learning-based computer-aided diagnosis (DL-CAD) system for detecting and characterizing lung nodules: comparison with the performance of double reading by radiologists. Thorac. Cancer10, 183–192 (2019). ArticleCASPubMedGoogle Scholar
- Majkowska, A. et al. Chest radiograph interpretation with deep learning models: assessment with radiologist-adjudicated reference standards and population-adjusted evaluation. Radiology294, 421–431 (2019). ArticlePubMedGoogle Scholar
- Park, S. et al. Deep learning-based detection system for multiclass lesions on chest radiographs: comparison with observer readings. Eur. Radiol.30, 1359–1368 (2019). ArticlePubMedGoogle Scholar
- Patel, B. N. et al. Human–machine partnership with artificial intelligence for chest radiograph diagnosis. npj Digital Med.2, 111 (2019). ArticleGoogle Scholar
- Rajpurkar, P. et al. Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLOS Med.15, e1002686 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Singh, R. et al. Deep learning in chest radiography: detection of findings and presence of change. PLoS ONE13, e0204155 (2018). ArticlePubMedPubMed CentralCASGoogle Scholar
- Walsh, S. L. F., Calandriello, L., Silva, M. & Sverzellati, N. Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: a case-cohort study. Lancet Respir. Med.6, 837–845 (2018). ArticlePubMedGoogle Scholar
- Wang, S. et al. 3D convolutional neural network for differentiating pre-invasive lesions from invasive adenocarcinomas appearing as ground-glass nodules with diameters ≤3 cm using HRCT. Quant. Imaging Med. Surg.8, 491–499 (2018). ArticleCASPubMedPubMed CentralGoogle Scholar
- Park, S. et al. Application of deep learning-based computer-aided detection system: detecting pneumothorax on chest radiograph after biopsy. Eur. Radio.29, 5341–5348 (2019). ArticleGoogle Scholar
- Lakhani, P. & Sundaram, B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology284, 574–582 (2017). ArticlePubMedGoogle Scholar
- Becker, A. S. et al. Deep learning in mammography: diagnostic accuracy of a multipurpose image analysis software in the detection of breast cancer. Investig. Radio.52, 434–440 (2017). ArticleGoogle Scholar
- Ciritsis, A. et al. Automatic classification of ultrasound breast lesions using a deep convolutional neural network mimicking human decision-making. Eur. Radio.29, 5458–5468 (2019). ArticleGoogle Scholar
- Cogan, T., Cogan, M. & Tamil, L. RAMS: remote and automatic mammogram screening. Comput. Biol. Med.107, 18–29 (2019). ArticlePubMedGoogle Scholar
- McKinney, S. M. et al. International evaluation of an AI system for breast cancer screening. Nature577, 89–94 (2020). ArticleCASPubMedGoogle Scholar
- Peng, W., Mayorga, R. V. & Hussein, E. M. A. An automated confirmatory system for analysis of mammograms. Comput. Methods Prog. Biomed.125, 134–144 (2016). ArticleCASGoogle Scholar
- Ribli, D., Horváth, A., Unger, Z., Pollner, P. & Csabai, I. Detecting and classifying lesions in mammograms with deep learning. Sci. Rep.8, 4165 (2018). ArticlePubMedPubMed CentralCASGoogle Scholar
- Rodríguez-Ruiz, A. et al. Detection of Breast cancer with mammography: effect of an artificial intelligence support system. Radiology290, 305–314 (2018). ArticlePubMedGoogle Scholar
- Rodriguez-Ruiz, A. et al. Stand-alone artificial intelligence for breast cancer detection in mammography: comparison with 101 radiologists. J. Natl Cancer Inst.111, 916–922 (2019). ArticlePubMedPubMed CentralGoogle Scholar
- Byra, M. et al. Breast mass classification in sonography with transfer learning using a deep convolutional neural network and color conversion. Med. Phys.46, 746–755 (2019). ArticlePubMedGoogle Scholar
- Choi, J. S. et al. Effect of a deep learning framework-based computer-aided diagnosis system on the diagnostic performance of radiologists in differentiating between malignant and benign masses on breast ultrasonography. Korean J. Radio.20, 749–758 (2019). ArticleGoogle Scholar
- Dalmis, M. U. et al. Artificial intelligence–based classification of breast lesions imaged with a multiparametric breast mri protocol with ultrafast DCE-MRI, T2, and DWI. Investig. Radiol.54, 325–332 (2019). ArticleGoogle Scholar
- Fujioka, T. et al. Distinction between benign and malignant breast masses at breast ultrasound using deep learning method with convolutional neural network. Jpn J. Radio.37, 466–472 (2019). ArticleGoogle Scholar
- Kim, S. M. et al. A comparison of logistic regression analysis and an artificial neural network using the BI-RADS Lexicon for ultrasonography in conjunction with introbserver variability. J. Digital Imaging25, 599–606 (2012). ArticleGoogle Scholar
- Truhn, D. et al. Radiomic versus convolutional neural networks analysis for classification of contrast-enhancing lesions at multiparametric breast MRI. Radiology290, 290–297 (2019). ArticlePubMedGoogle Scholar
- Wu, N. et al. Deep neural networks improve radiologists’ performance in breast cancer screening. IEEE Trans. Med. Imaging39, 1184–1194 (2020). ArticlePubMedGoogle Scholar
- Yala, A., Schuster, T., Miles, R., Barzilay, R. & Lehman, C. A deep learning model to triage screening mammograms: a simulation study. Radiology293, 38–46 (2019). ArticlePubMedGoogle Scholar
- Zhou, J. et al. Weakly supervised 3D deep learning for breast cancer classification and localization of the lesions in MR images. J. Magn. Reson. Imaging50, 1144–1151 (2019). ArticlePubMedGoogle Scholar
- Li, Z. et al. Efficacy of a deep learning system for detecting glaucomatous optic neuropathy based on color fundus photographs. Ophthalmology125, 1199–1206 (2018). ArticlePubMedGoogle Scholar
- Lin, H. et al. Diagnostic efficacy and therapeutic decision-making capacity of an artificial intelligence platform for childhood cataracts in eye clinics: a multicentre randomized controlled trial. EClinicalMedicine9, 52–59 (2019). ArticlePubMedPubMed CentralGoogle Scholar
- Annarumma, M. et al. Automated triaging of adult chest radiographs with deep artificial neural networks. Radiology291, 196–202 (2019). ArticlePubMedGoogle Scholar
- Yala, A., Lehman, C., Schuster, T., Portnoi, T. & Barzilay, R. A deep learning mammography-based model for improved breast cancer risk prediction. Radiology292, 60–66 (2019). ArticlePubMedGoogle Scholar
- Sedgwick, P. Meta-analyses: how to read a funnel plot. BMJ346, f1342 (2013). ArticleGoogle Scholar
- Herent, P. et al. Detection and characterization of MRI breast lesions using deep learning. Diagn. Inter. Imaging100, 219–225 (2019). ArticleCASGoogle Scholar
- Bossuyt, P. M. et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ351, h5527 (2015). ArticlePubMedPubMed CentralGoogle Scholar
- Sounderajah, V. et al. Developing specific reporting guidelines for diagnostic accuracy studies assessing AI interventions: the STARD-AI Steering Group. Nat. Med.26, 807–808 (2020). ArticleCASPubMedGoogle Scholar
- Vollmer, S. et al. Machine learning and artificial intelligence research for patient benefit: 20 critical questions on transparency, replicability, ethics, and effectiveness. BMJ368, l6927 (2020). ArticlePubMedGoogle Scholar
- Liu, X. et al. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat. Med.26, 1364–1374 (2020). ArticleCASPubMedPubMed CentralGoogle Scholar
- Whiting, P. F. et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med.155, 529–536 (2011). ArticlePubMedGoogle Scholar
- Food, U. & Administration, D. Artificial Intelligence and Machine Learning in Software as a Medical Device (US Food and Drug Administratio, 2019).
- Titano, J. J. et al. Automated deep-neural-network surveillance of cranial images for acute neurologic events. Nat. Med.24, 1337–1341 (2018). ArticleCASPubMedGoogle Scholar
- Rankin, D. et al. Reliability of supervised machine learning using synthetic data in health care: Model to preserve privacy for data sharing. JMIR Med. Inform.8, e18910 (2020). ArticlePubMedPubMed CentralGoogle Scholar
- Cawley, G. C. & Talbot, N. L. On over-fitting in model selection and subsequent selection bias in performance evaluation. J. Mach. Learn. Res.11, 2079–2107 (2010). Google Scholar
- Blalock, D., Ortiz, J., Frankle, J. & Guttag, J. What is the state of neural network pruning? Preprint at https://arxiv.org/abs/2003.03033 (2020).
- Beam, A. L., Manrai, A. K. & Ghassemi, M. Challenges to the reproducibility of machine learning models in health care. JAMA323, 305–306 (2020). ArticlePubMedPubMed CentralGoogle Scholar
- Celi, L. A. et al. Bridging the health data divide. J. Med. Internet Res.18, e325 (2016). ArticlePubMedPubMed CentralGoogle Scholar
- Shah, P. et al. Artificial intelligence and machine learning in clinical development: a translational perspective. npj Digital Med.2, 69 (2019). ArticleGoogle Scholar
- Collins, G. S., Reitsma, J. B., Altman, D. G. & Moons, K. G. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ350, g7594 (2015). ArticlePubMedGoogle Scholar
- Schulz, K. F., Altman, D. G. & Moher, D. CONSORT 2010 Statement: updated guidelines for reporting parallel group randomised trials. BMJ340, c332 (2010). ArticlePubMedPubMed CentralGoogle Scholar
- Chan, A.-W. et al. SPIRIT 2013 statement: defining standard protocol items for clinical trials. Ann. Intern. Med.158, 200–207 (2013). ArticlePubMedPubMed CentralGoogle Scholar
- Cruz Rivera, S. et al. Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension. Nat. Med.26, 1351–1363 (2020). ArticleCASPubMedPubMed CentralGoogle Scholar
- Collins, G. S. & Moons, K. G. Reporting of artificial intelligence prediction models. Lancet393, 1577–1579 (2019). ArticlePubMedGoogle Scholar
- Ngiam, K. Y. & Khor, I. W. Big data and machine learning algorithms for health-care delivery. Lancet Oncol.20, e262–e273 (2019). ArticlePubMedGoogle Scholar
- World Health Organization. Big Data and Artificial Intelligence for Achieving Universal Health Coverage: an International Consultation on Ethics: Meeting Report, 12–13 October 2017 (World Health Organization, 2018).
- Cath, C., Wachter, S., Mittelstadt, B., Taddeo, M. & Floridi, L. Artificial Intelligence and the ‘Good Society’: the US, EU, and UK approach. Sci. Eng. Ethics24, 505–528 (2018). PubMedGoogle Scholar
- Mittelstadt, B. The ethics of biomedical ‘Big Data’ analytics. Philos. Technol.32, 17–21 (2019). ArticleGoogle Scholar
- McInnes, M. D. F. et al. Preferred reporting items for a systematic review and meta-analysis of diagnostic test accuracy studies: the PRISMA-DTA statement. JAMA319, 388–396 (2018). ArticlePubMedGoogle Scholar
- Reitsma, J. B. et al. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J. Clin. Epidemiol.58, 982–990 (2005). ArticlePubMedGoogle Scholar
- DerSimonian, R. & Laird, N. Meta-analysis in clinical trials. Controlled Clin. Trials7, 177–188 (1986). ArticleCASPubMedGoogle Scholar
- Jones, C. M., Ashrafian, H., Darzi, A. & Athanasiou, T. Guidelines for diagnostic tests and diagnostic accuracy in surgical research. J. Investig. Surg.23, 57–65 (2010). ArticleGoogle Scholar
Acknowledgements
Infrastructure support for this research was provided by the NIHR Imperial Biomedical Research Centre (BRC).






